Introduction to Gaussian Processes for Time Dependent Data
VectorByte Methods Training
Gaussian Process: Introduction
A Gaussian Process model is a non paramteric and flexible regression model.
It started being used in the field of spatial statistics, where it is called kriging.
It is also widely used in the field of machine learning since it makes fast predictions and gives good uncertainty quantification commonly used as a surrogate model. (Gramacy 2020)
Uses and Benefits
Surrogate Models: Imagine a case where field experiments are infeasible and computer experiments take a long time to run, we can approximate the computer experiments using a surrogate model.
It can be used to calibrate computer models w.r.t the field observations or computer simulations.
The ability to this model to provide good UQ makes it very useful in other fields such as ecology where the data is sparse and noisy and therefore good uncertainty measures are paramount.
What is a GP?
Here, we assume that the data come from a Multivariate Normal Distribution (MVN).
We then make predictions conditional on the data.
We are essentially taking a “fancy” average of the data to make predictions
To understand how it works, let’s first visualize this concept and then look into the math
Visualizing a GP
How does a GP work
Our “fancy” average was just using data around the new location.
We are using points closer to each other to correlate the responses.
Now we know, we are averaging the data “nearby”… How do you define that?
- This indicates that we are using distance in some way. Where…? We need some math
Mathematically
Any normal distribution can be described by a mean vector \mu and a covariance matrix \Sigma.
Mathematically, we can write it as,
Y_{\ n \times 1} \sim N \ ( \ \mu(X)_{\ n \times 1} \ , \ \Sigma(X)_{ \ n \times n} \ ) Here, Y is the response of interest and n is the number of observations.
- Our goal is to find Y_p \ \vert \ Y, X.
Distance
- Recall that in Linear Regression, you have \ \Sigma \ = \sigma^2 \mathbb{I}
For a GP, the covariance matrix ( \Sigma ) is defined by a kernel.
Consider,
\Sigma_n = \tau^2 C_n where C_n = \exp \left( - \vert \vert x - x' \vert \vert^2 \right), and x and x' are input locations.
Interpretation of the kernel
C_n = \exp \left( - \vert \vert x - x' \vert \vert^2 \right)
The covariance structure now depends on how close together the inputs. If inputs are close in distance, then the responses are more highly correlated.
The covariance will decay at an exponential rate as x moves away from x'.
Summarizing our data
Now we will learn how to use a GP to make predictions at new locations.
As we learnt, we condition on the data. We can think of this as the prior.
Now, consider, (\mathcal{X}, \mathcal{Y}) as the predictive set.
How to make predictions
The goal is to find the distribution of \mathcal{Y} \ \vert X_n, Y_n which in this case is the posterior distribution.
By properties of Normal distribution, the posterior is also normally distributed.
We also need to write down the mean and variance of the posterior distribution so it’s ready for use.
How to make predictions
- First we will “stack” the predictions and the data.
- Now, let’s denote the predictive mean with \mu(\mathcal{X}) and predictive variance with \sigma^2(\mathcal{X})
Distribution of Interest!
- We will apply the properties of conditional Normal distributions.
- Everything we do, relies on these equations. Now, let’s learn more about \Sigma
Sigma Matrix
- \Sigma_n = \tau^2 C_n where C_n is our kernel.
- One of the most common kernels which we will focus on is the squared exponential distance kernel written as
C_n = \exp{ \left( -\frac{\vert\vert x - x' \vert \vert ^2}{\theta} \right ) + g \mathbb{I_n}}
- What’s \tau^2, g and \theta though? No more math. We will just conceptually go through these
Hyper Parameters
A GP is non parameteric, however, has some hyper-parameters. In this case,
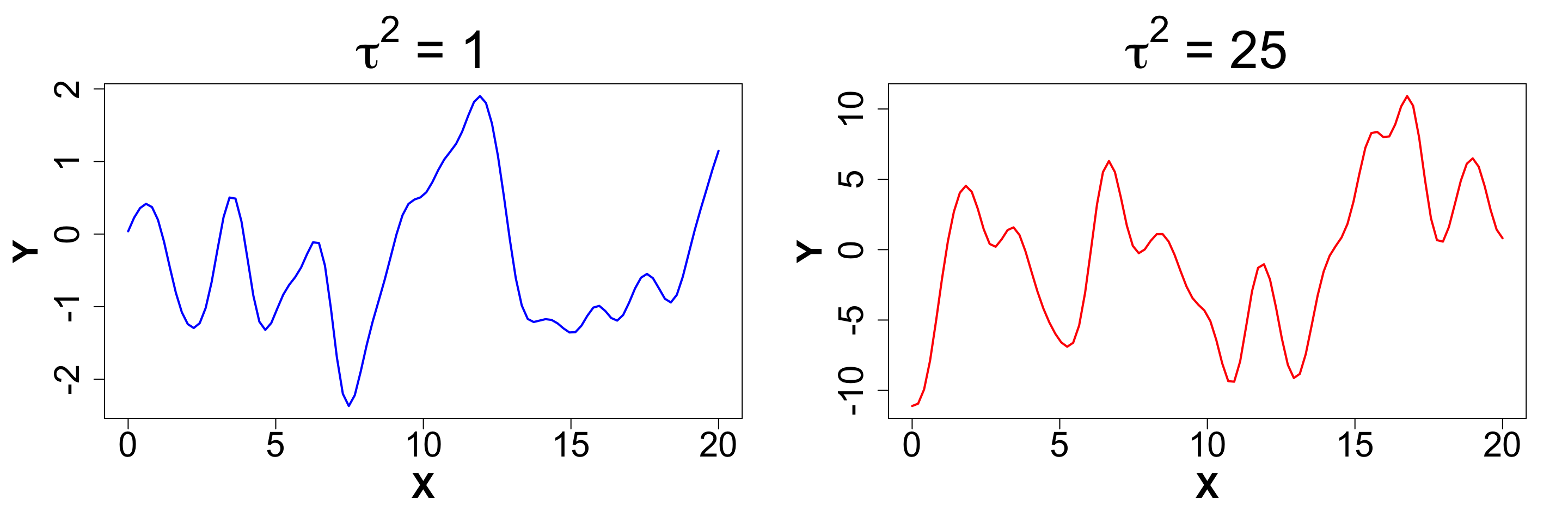
\tau^2 (Scale): This parameter can be used to adjust the amplitude of the data.
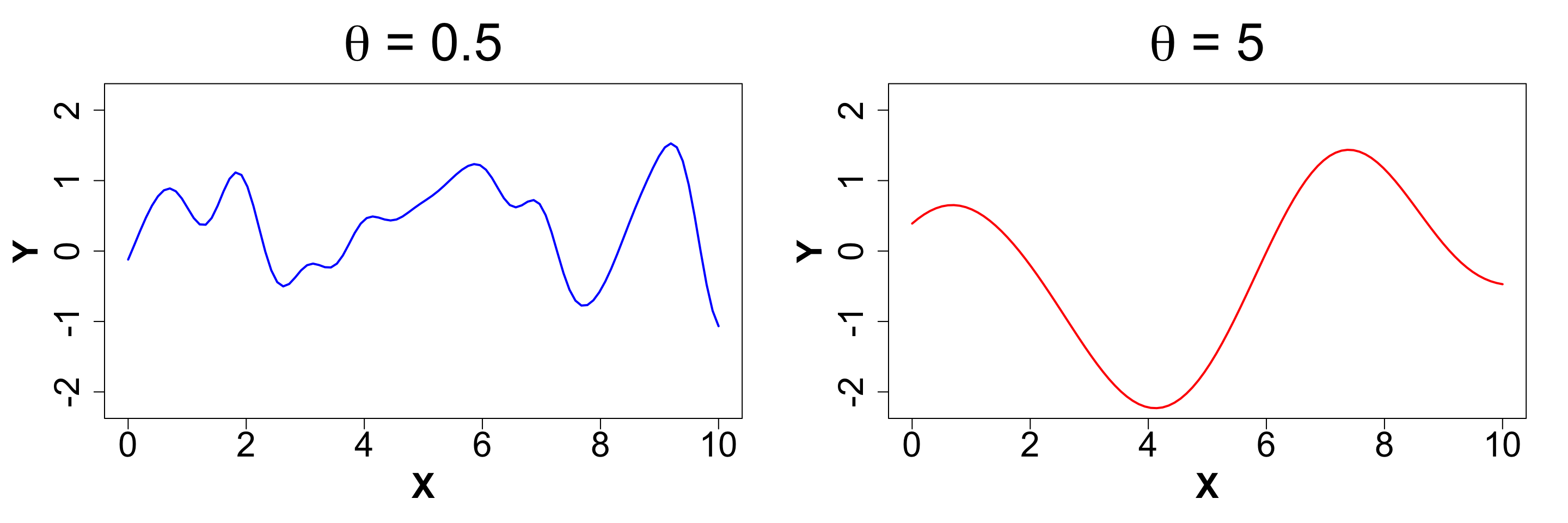
\theta (Length-scale): This parameter controls the rate of decay of correlation.
g (Nugget): This parameter controls the noise in the covariance structure (adds discontinuity)
Scale (Amplitude)
A random draw from a multivariate normal distribution with \tau^2 = 1 will produce data between -2 and 2.
Now let’s visualize what happens when we increase \tau^2 to 25.
Length-scale (Rate of decay of correlation)
Determines how “wiggly” a function is
Smaller \theta means wigglier functions i.e. visually:
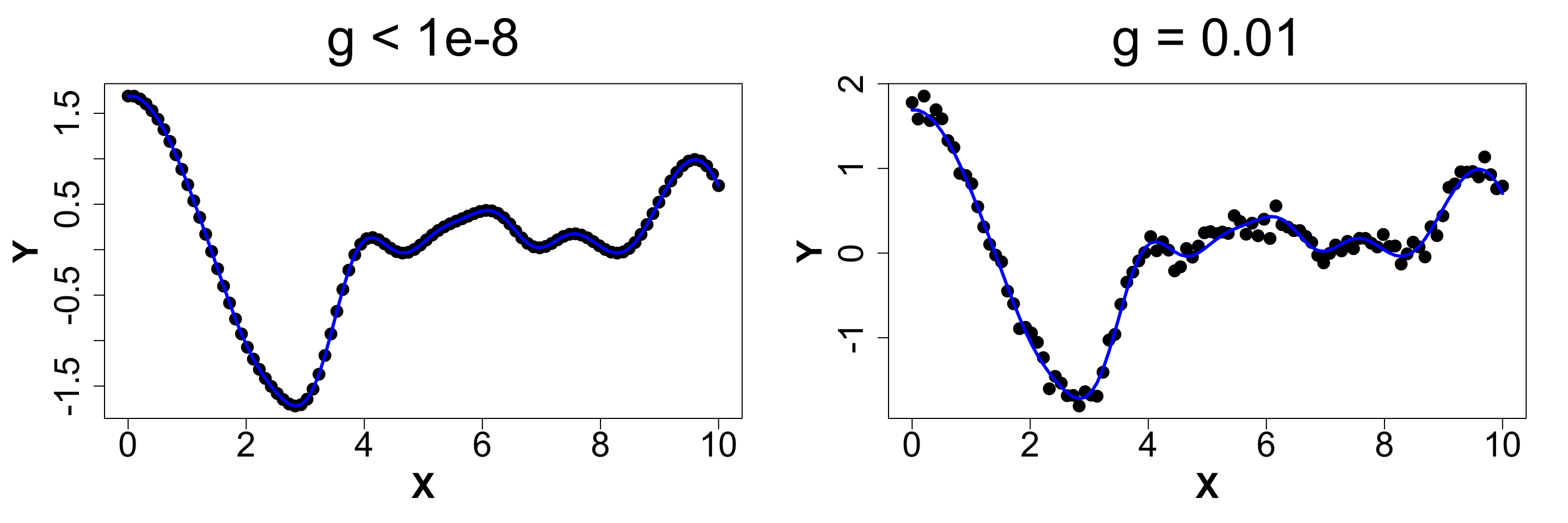
Nugget (Noise)
Ensures discontinuity and prevents interpolation which in turn yields better UQ.
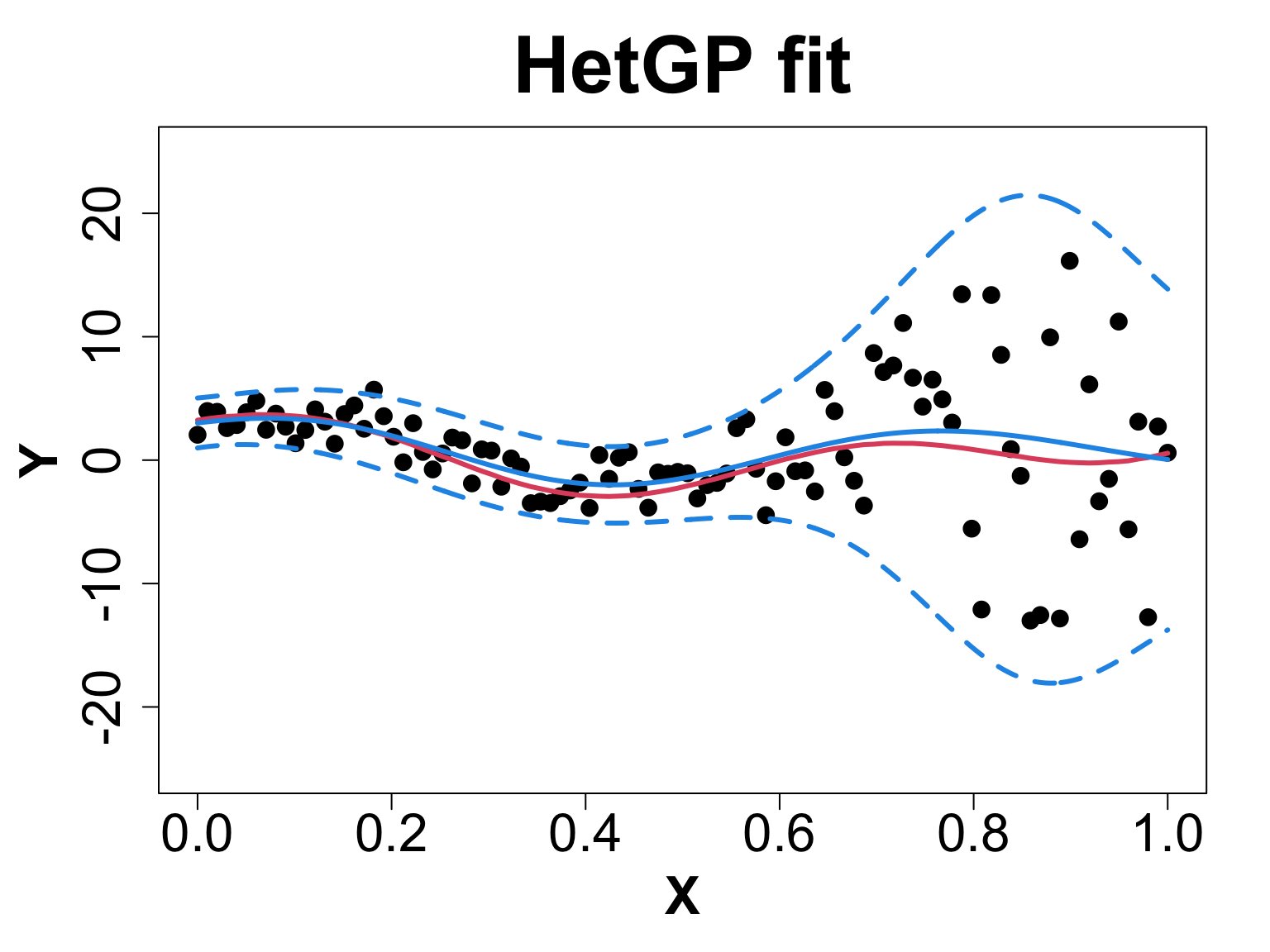
We will compare a sample from g ~ 0 (< 1e-8 for numeric stability) vs g = 0.1 to observe what actually happens.
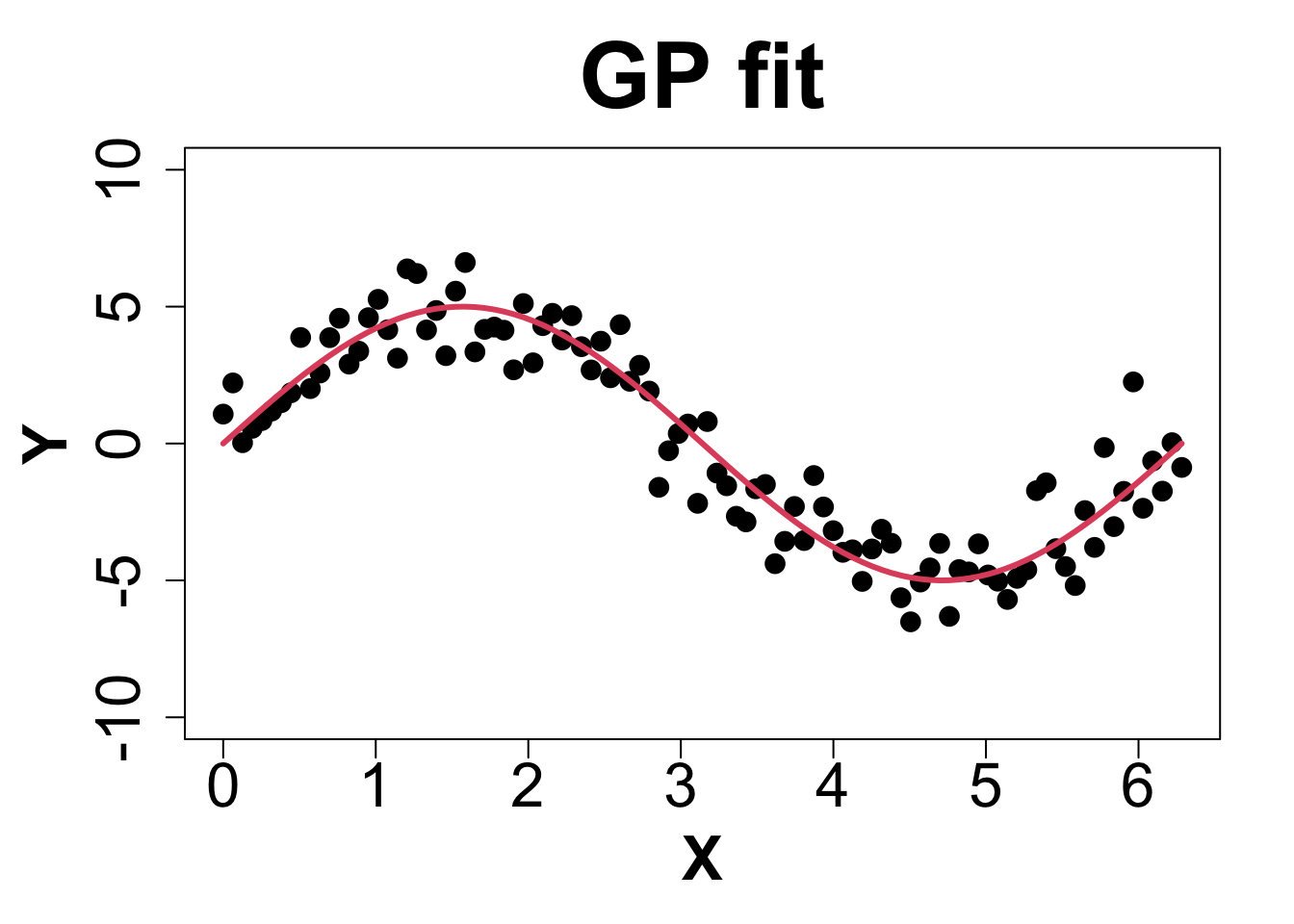
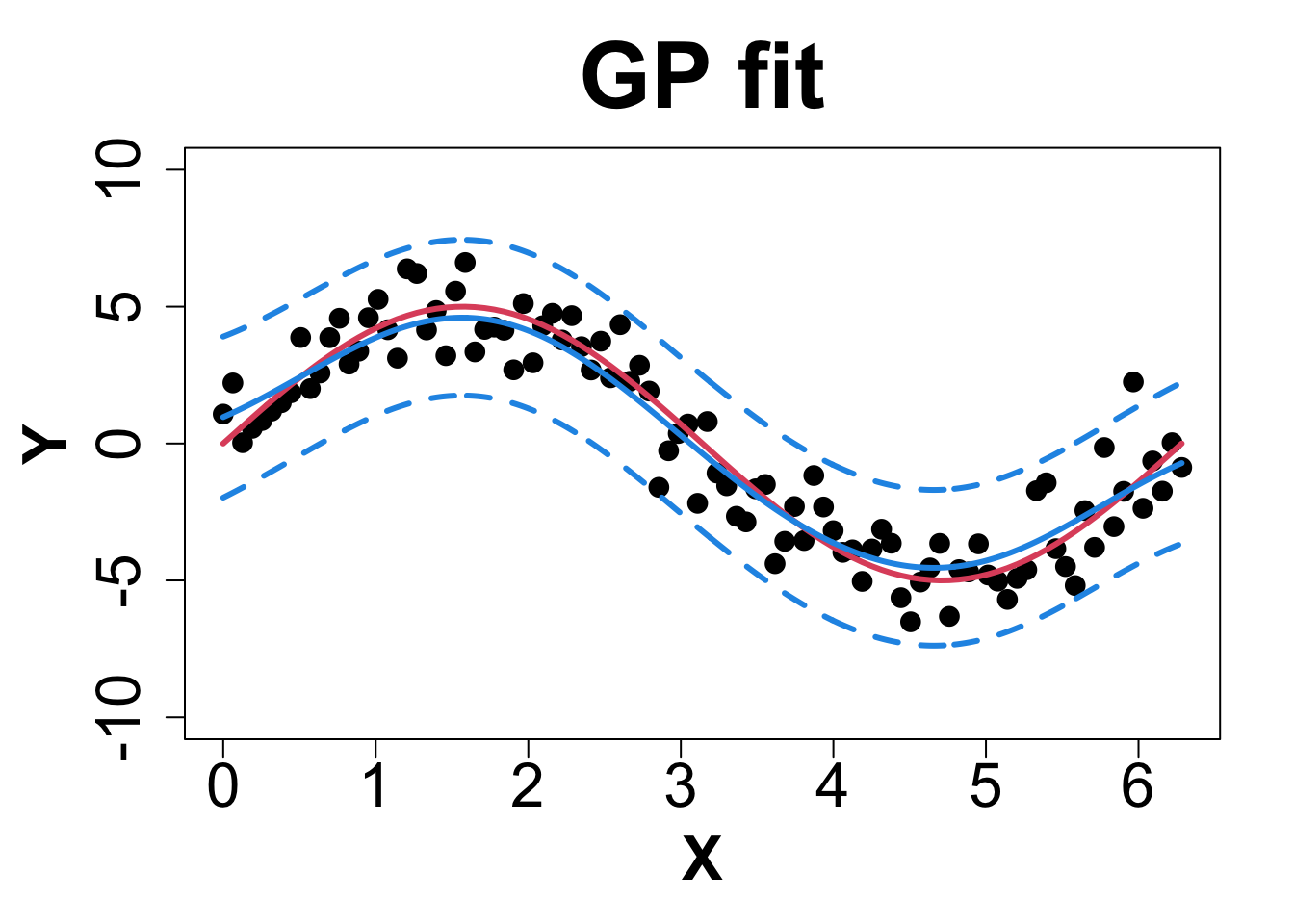
Toy Example (1D Example)
Toy Example (1D Example)
Extentions
Anisotropic Gaussian Processes: Suppose our data is multi-dimensional, we can control the length-scale (\theta) for each dimension.
Heteroskedastic Gaussian Processes: Suppose our data is noisy and the noise is input dependent, then we can use a different nugget for each unique input rather than a scalar g.
Anisotropic Gaussian Processes
In this situation, we can rewrite the C_n matrix as,
C_\theta(x , x') = \exp{ \left( -\sum_{k=1}^{m} \frac{ (x_k - x_k')^2 }{\theta_k} \right ) + g \mathbb{I_n}}
Here, \theta = (\theta_1, \theta_2, …, \theta_m) is a vector of length-scales, where m is the dimension of the input space.
Heteroskedastic Gaussian Processes
- Heteroskedasticity implies that the data is noisy, and the noise is input dependent and irregular. (Binois, Gramacy, and Ludkovski 2018)
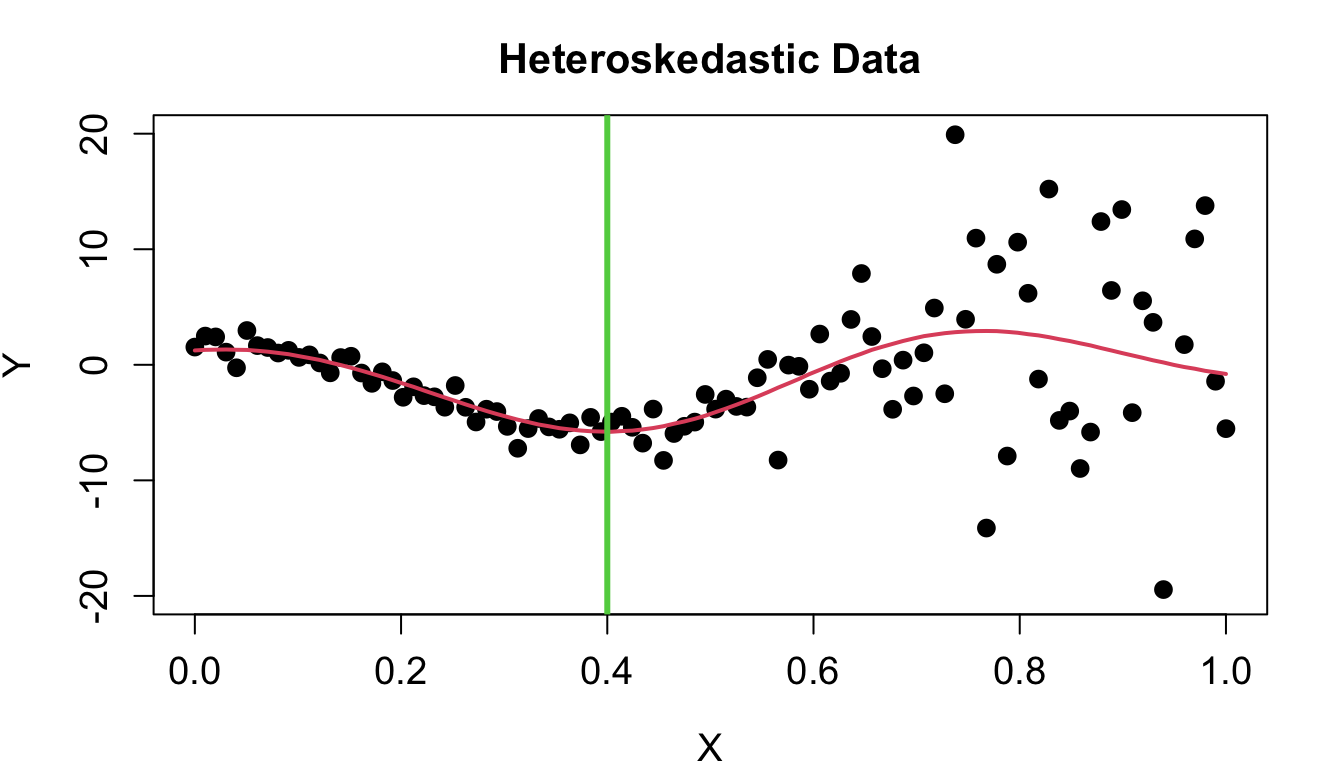
HetGP Setup
Let Y_n be the response vector of size n.
Let X = (X_1, X_2 ... X_n) be the input space.
Then, a regular GP is written as:
\begin{align*} Y_N \ & \ \sim GP \left( 0 \ , \tau^2 C_n \right); \ \text{where, }\\[2pt] C_n & \ = \exp{ \left( -\frac{\vert\vert x - x' \vert \vert ^2}{\theta} \right ) + g \mathbb{I_n}} \end{align*}
HetGP Setup
In case of a hetGP, we have:
\begin{aligned} Y_n\ & \ \sim GP \left( 0 \ , \tau^2 C_{n, \Lambda} \right) \ \ \text{where, }\\[2pt] C_{n, \Lambda} & \ = \exp{ \left( -\frac{\vert\vert x - x' \vert \vert ^2}{\theta} \right ) + \Lambda_n} \ \ \ \text{and, }\ \ \\[2pt] \ \ \Lambda_n \ & \ = \ \text{Diag}(\lambda_1, \lambda_2 ... , \lambda_n) \\[2pt] \end{aligned}
- Instead of one nugget for the GP, we have a vector of nuggets i.e. a unique nugget for each unique input.
HetGP Predictions
- Recall, for a GP, we make predictions using the following:
- We can make predictions using these same equations with \Sigma(X_n) \ \ = \tau^2 C_{n, \Lambda}
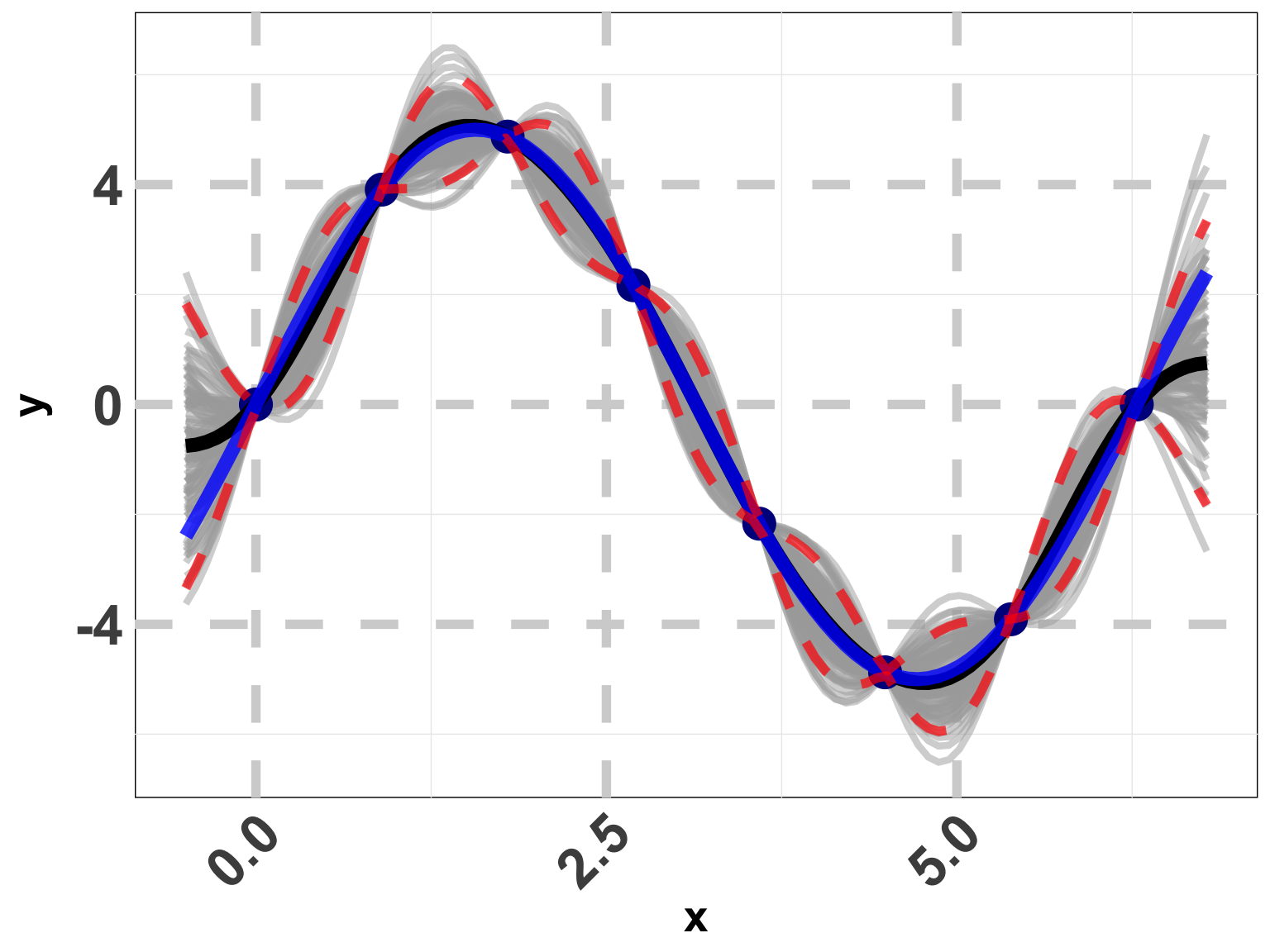
Toy Example (1D Example)
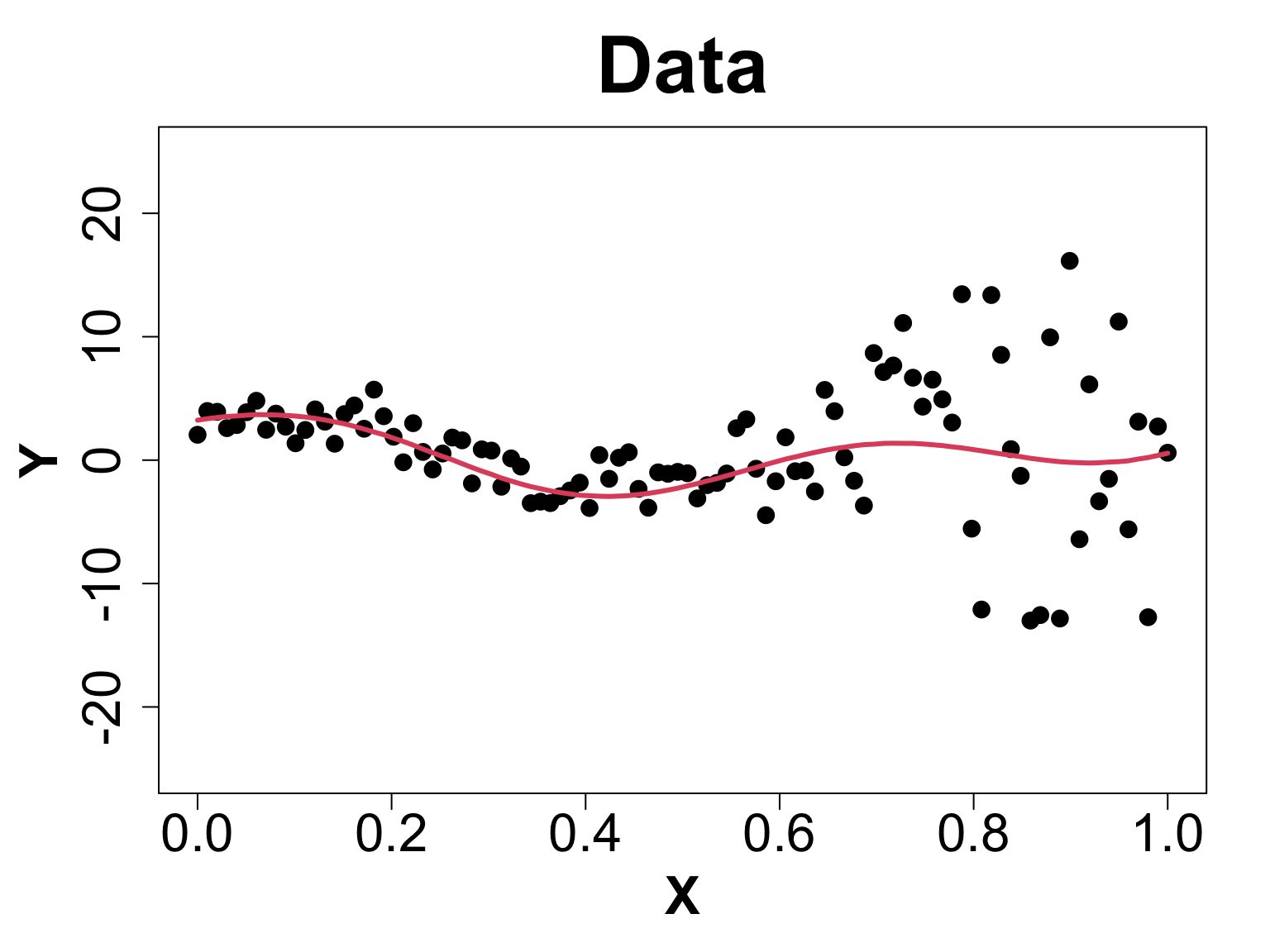
Toy Example (1D Example)
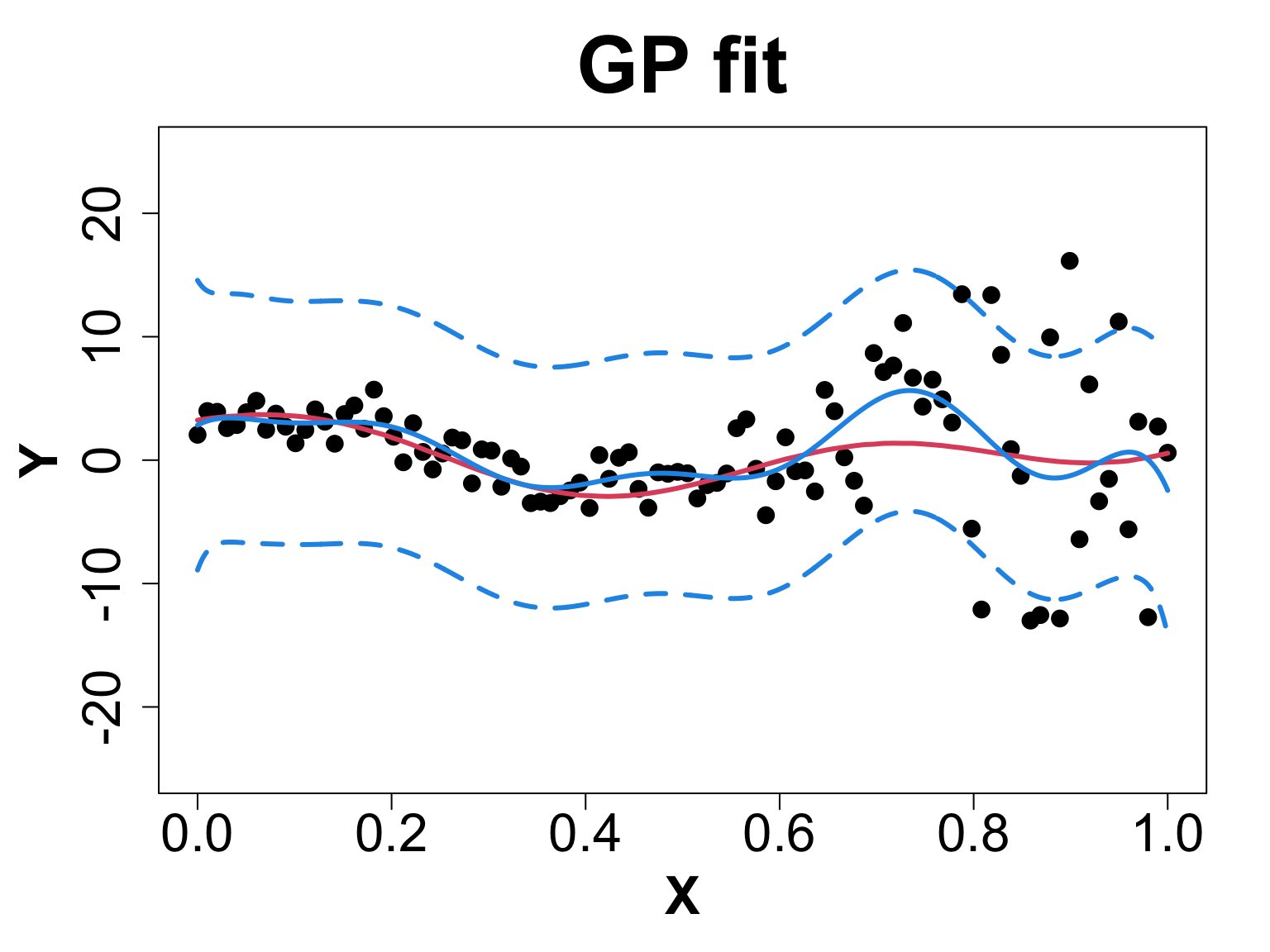
Toy Example (1D Example)
Intro to Ticks Problem
EFI-RCN held an ecological forecasting challenge NEON Forecasting Challenge (Thomas et al. 2022)
We focus on the Tick Populations theme which studies the abundance of the lone star tick (Amblyomma americanum)
Tick Population Forecasting
Some details about the challenge:
Objective: Forecast tick density for 4 weeks into the future
Sites: The data is collected across 9 different sites, each plot was of size 1600m^2 using a drag cloth
Data: Sparse and irregularly spaced. We only have ~650 observations across 10 years at 9 locations
Predictors
X_1 Iso-week: The week in which the tick density was recorded.
X_2 Sine wave: \left( \text{sin} \ ( \frac{2 \ \pi \ X_1}{106} ) \right)^2.
X_3 Greenness: Environmental predictor (in practical)
Practical
- Setup these predictors
- Transform the data to normal
- Fit a GP to the Data
- Make Predictions on a testing set
- Check how predictions perform.